Hi @MathieuBourdenx
The difference you are observing is due to a change we made in the default configuration parameters with which we run the MapMyCells algorithm (see this page and this webinar at around the 14 minute mark for a description of the MapMyCells algorithm).
At each level in the taxonomy, the bootstrapping_probability is determined as follows: we map each cell bootstrap_iteration times (bootstrap_iteration is a configurable integer >= 1), with each iteration using a random subset of bootstrap_factor of the whole set of marker genes (bootstrap_factor is a configurable parameter between that is > 0 and <= 1). We then choose the cell type that is selected by the plurality of the independent mapping iterations. The bootstrapping_probability is the fraction of iterations that chose the selected cell type. Ideally, low quality cells will have a low bootstrapping_probability, since the random subsampling of marker genes will result in a diversity of cell type assignments across iterations. High quality cells will have a high bootstrapping_probability, because data is so unambiguous that the change in marker gene subset will have little effect on the result of mapping.
Historically, we have run the algorithm with
bootstrap_iteration = 100
bootstrap_factor = 0.9
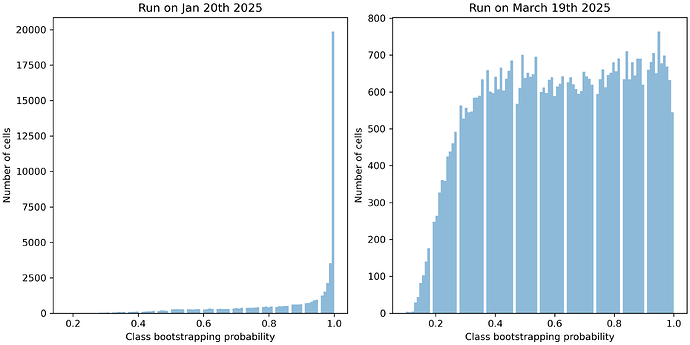
We have found that randomly subsetting 90% of the marker genes at each iteration did not yield a helpful value of bootstrapping_probability. 90% was so high that even low quality cells had a fairly consistent mapping and every cell had bootstrapping_probability > 0.9, regardless of quality.
Given that, we have adjusted the algorithm to use
bootstrap_iteraiton = 100
bootstrap_factor = 0.5
Now, each independent iteration of mapping has a more diverse set of marker genes, causing low quality cells to actually have a low bootstrapping_probability, an indication that the mapping may be suspect (depending on your tolerance for certainty of mapping).
For reference, you should be able to see these changes in configuration if you load the JSON output file and look at the metadata stored under the config key.
To offer a more concrete justification for this change:
The bootstrapping_probability at each level in the taxonomy can be thought of as a conditional probability (“the probability that the assignment that this level is correct, assuming that the assignments at the higher levels of the taxonomy are correct”). To get an unconditional probability, you can just take a running product of the probabilities starting at the highest level of the taxonomy (this is reported as the “aggregate_probability” in the JSON result file). With the change from bootstrap_factor=0.9 to bootstrap_factor=0.5, we found that the aggregate probability became a much better predictor of a mapping’s accuracy. When we performed a test-train hold out assessment using the Yao et al. transcriptomics data, we found that, for instance, if we took all the cells with aggregate_probability=0.6, roughly 60% of them were correct and 40% of them were incorrect (likewise, 20% of assignments with aggregate_probability=0.8 were incorrect). With bootstrap_factor=0.9, again, nearly every cell at every level had bootstrapping_probability > 0.9, regardless of whether or not the mapping was correct, so aggregate_probability really didn’t provide an easily interpretable metric for the quality of the mapping.
In our test-train evaluation, the change from bootstrap_factor=0.9 to bootstrap_factor=0.5 had almost no effect on the fraction of cells assigned to the correct cell type so, according to our tests, the only effect was that aggregate_probability became a better metric of quality.
So, in summary: while the bootstrapping_probability for your January run looked very good, that was deceptive. bootstrap_factor=0.9 is so large that even cells with a poor quality mapping received bootstrapping_probability ~ 1.0. I suspect that the new results actually reflect the quality of mapping for your cells.
Please let me know if you have any other questions or if anything is unclear.