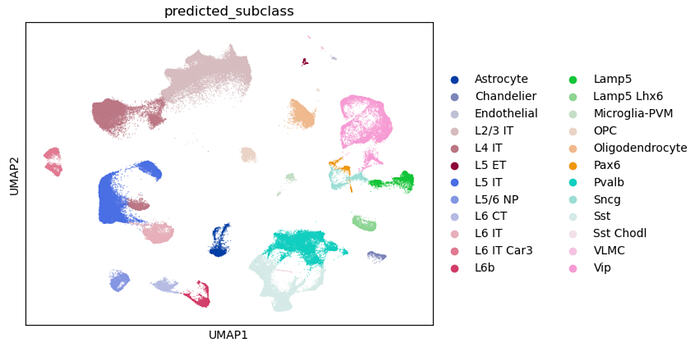
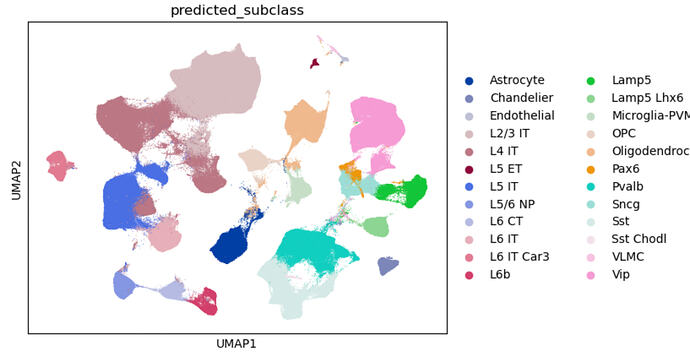
I created a query data set using SEA-AD snRNAseq MTG data from the raw fastqs at sage Synape (with clinical consensus diagnosis of Alzheimers disease and Control). I ran cell ranger (with introns) and perfomed QC. The ref I am using the MTG final_nuclei ref provided in the AWS registry by Gabitto et al.
I have large portion of the query that wont match the ref.
Top (Just Reference)
Bottom (Reference + Query)
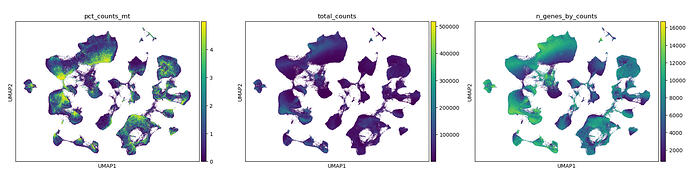
I used cellbender to perform ambient RNA correction and have done every QC I think of imaginable. I don’t understand what this cell state is and why its not mapping well to the ref.
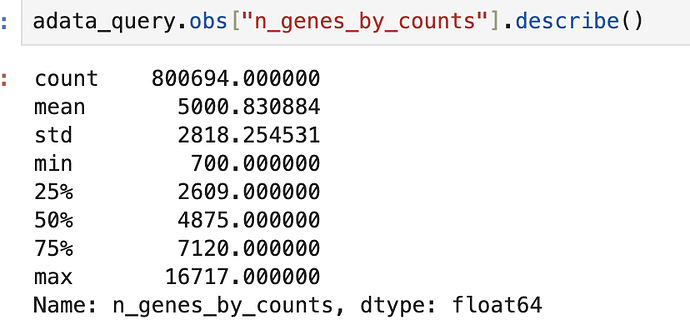
Please look at my QC below.
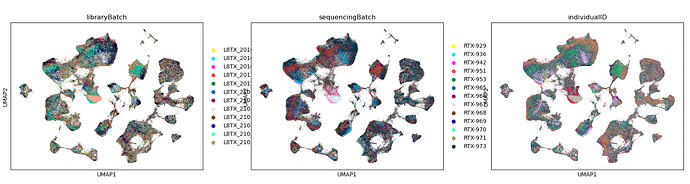
Also I think it might be related donor effects by am I not sure :
Did you encounter something like this, if so how did you deal with it?
Also did you all use any ambient correction method if so which one?